There are many functions that read/write data in R and export it files
read.table, read.csv for reading tabular data and return it in data frames from delimited files. read.csv is identical to read.table except that default separator is coma. There are so many arguments for these functions, the most commonly used arguments are:
- file : string –> file name or URL
- header : logical –> TRUE if the file first line is a header line, FALSE if first line is data.
- sep : string –> represents the data separator
- quote “ string –> if character values are enclosed in quotes, this argument should specify the type of quotes.
- colClasses : string vector –> represent the data tyoe of each column in file.
- nrows : integer –> the number of rows in file (optional)
- comment.char : string –> the comment indicator string
- skip : integer –> number of lines to skip from the file beginning
- stringsAsFactors : logical –> should character variables be coded as factors?
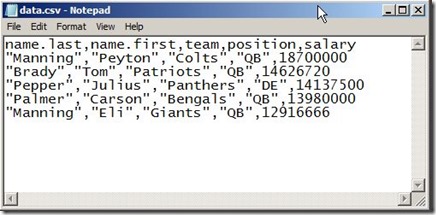
save the following file into your default user directory
now you can load it into R
You may process this data and want to save it again in another file. To save data as text file, use write.table which have many parameters similar to read.table. There are many wrapper functions that call write.table with different defaults like write.csv()
readLines and writeLines can be used to read/write certain number of lines of a text file.
There are more read/write functions that are available in R but we introduced here only the most commonly used ones.
Get/Set your working directory
You may be wondering how R knows the path to data.csv file ? R look for files in the user working directory for files and this working directory by default is your documents folder (that’s why we save the file in “My Documents” in the first place"). To know you working directory, use getwd()
to change your working directory, go to File >> Change dir and select your new working directory and click Ok.
You can use dir() to list all files in your working directory.
Stay tuned for more R notes.